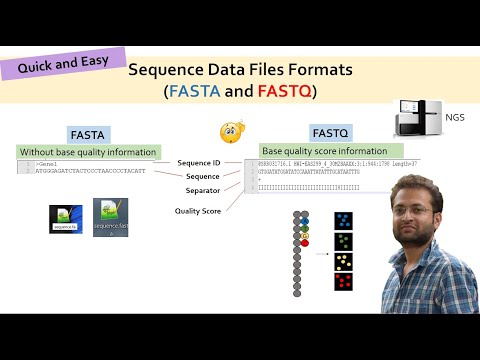
ADS1: Reads in FASTQ format
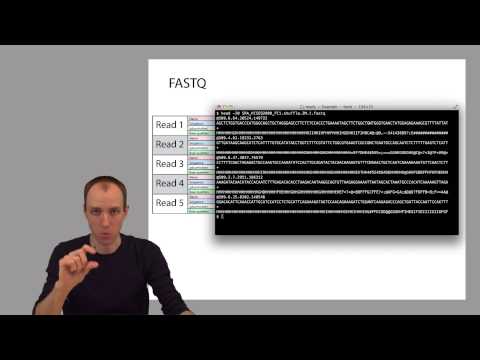
ADS1: Practical: Working with sequencing readsПодробнее

Introducing FASTQ FilesПодробнее

ADS1: Practical: Matching real readsПодробнее
Bioinformatics Analysis of Quality Scores from FASTQ file using BiopythonПодробнее

STAT115 Chapter 3.2 FASTQ and FASTQCПодробнее

FASTQ FormatПодробнее

FastQ Sequence Data In BioconductorПодробнее

ADS1: Practical: Downloading and reading a genomeПодробнее

Bioinformatics Filtering FASTQ files by PHRED quality scores with BiopythonПодробнее

fastq, fasta, and gzipПодробнее

FASTQ file formatПодробнее

Section 4.1 read FASTQ fileПодробнее

Processing raw sequencing reads, Elena GhibanПодробнее

Bioinformatics - fastp FastQ Preprocessing Tool (Timestamps)Подробнее

Fasta and Fastq Files Session3.2Подробнее

Bioinformatics_101: How to count read number in fastq fileПодробнее

Difference between FASTA and FASTQ file formats. How to make a fasta formatted file.Подробнее