Degraded FFPE-RNA Samples Sequencing and Analysis | Protocol Preview

Analysis of FFPE tumor samplesПодробнее

Sequencing FFPE Samples: IntroductionПодробнее

FFPE Simplified: The nCounter Analysis System and Analyzing FFPE SamplesПодробнее
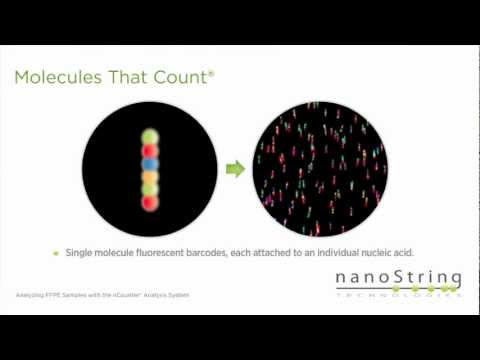
Expression profiling of archival FFPE samplesПодробнее

Unbiased Deep Sequencing of RNA Viruses from Clinical Samples | Protocol PreviewПодробнее

Retrospective MicroRNA Sequencing by FFPE RNA | Protocol PreviewПодробнее

Improved RNA-Seq Data Quality from Low-Input and FFPE Samples Using Enzyme-Free Ribosomal RNA...Подробнее

MERSCOPE Imaging in FFPE SamplesПодробнее

Considerations for RNA-sequencing with Degraded InputsПодробнее

Covaris Seminar: AFA® Technology for High Quality Next-Generation Sequencing Data in FFPE SamplesПодробнее

Visium for FFPE Gene Expression Protocol | Tissue Brightfield ImagingПодробнее

Unlocking FFPE tissues: The automation of library preparation of archival samples for RNA-SeqПодробнее

Webinar: RNA:Seq: Addressing the Challenges using KAPA RNA HyperPrepПодробнее

FFPE in your NGS StudyПодробнее
[COVARIS Webinar] AFA® Technology for High Quality Next-Generation Sequencing Data in FFPE SamplesПодробнее
![[COVARIS Webinar] AFA® Technology for High Quality Next-Generation Sequencing Data in FFPE Samples](https://img.youtube.com/vi/sfrd6ySmLxk/0.jpg)
Best Practices for Obtaining RNA Sequencing Data from Formalin-fixed Paraffin-embedded SamplesПодробнее

FFPE Simplified: The nCounter® Analysis System and FFPE SamplesПодробнее

USER TALK: Unlocking the Transcriptomic Potential of FFPE Cancer SamplesПодробнее

USER TALK: Reliable RNA-Seq Expression Profiling from Low-Quality FFPE Biobank SamplesПодробнее
